An interactive chemical viewer for 2D structures of small molecules
mols2grid
mols2grid is an interactive chemical viewer for 2D structures of small molecules, based on RDKit.
🐍 Installation
mols2grid was developped for Python 3.6+ and requires rdkit (>=2019.09.1), pandas and jinja2 as dependencies.
To install mols2grid from a clean conda environment:
conda install -c conda-forge 'rdkit>=2019.09.1'
pip install mols2grid
It is compatible with Jupyter Notebook and Google Colab (Visual Studio notebooks and Jupyterlab are not supported) and can run on Streamlit.
📜 Usage
import mols2grid
mols2grid.display("path/to/molecules.sdf",
# RDKit's MolDrawOptions parameters
fixedBondLength=25,
# rename fields for the output document
mapping={"SOL": "Solubility",
"SOL_classification": "Class",
"NAME": "Name"},
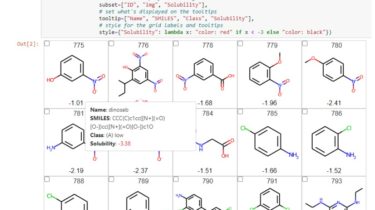
# set what's displayed on the grid
subset=["ID", "img", "Solubility"],
# set what's displayed on the tooltips
tooltip=["Name", "smiles", "Class", "Solubility"],
# style for the grid labels