Graphical tool to make photo collage posters in python
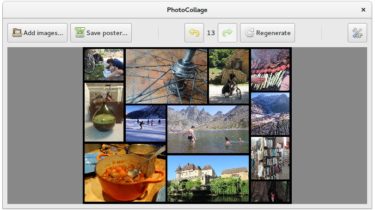
PhotoCollage Graphical tool to make photo collage posters PhotoCollage allows you to create photo collage posters. It assembles the input photographs it is given to generate a big poster. Photos are automatically arranged to fill the whole poster, then you can change the final layout, dimensions, border or swap photos in the generated grid. Eventually the final poster image can be saved in any size. The algorithm generates random layouts that place photos while taking advantage of all free space. […]
Read more