Cancer Drug Response Prediction via a Hybrid Graph Convolutional Network
DeepCDR
Cancer Drug Response Prediction via a Hybrid Graph Convolutional Network
This work has been accepted to ECCB2020 and was also published in the journal Bioinformatics.
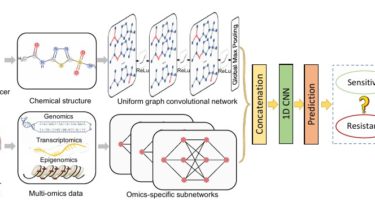
DeepCDR is a hybrid graph convolutional network for cancer drug response prediction. It takes both multi-omics data of cancer cell lines and drug structure as inputs and predicts the drug sensitivity (binary or contineous IC50 value).
- Keras==2.1.4
- TensorFlow==1.13.1
- hickle >= 2.1.0
DeepCDR can be downloaded by
git clone https://github.com/kimmo1019/DeepCDR
Installation has been tested in a Linux/MacOS platform.
We provide detailed step-by-step instructions for running DeepCDR model including data preprocessing, model training, and model test.
Model implementation
Step 1: Data Preparing
Three types of raw data are required to generate genomic mutation matrix, gene expression matrix and DNA methylation matrix from CCLE