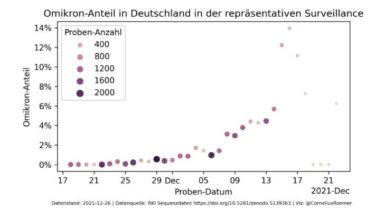
This repository contains a join of the metadata and pango lineage tables of all German SARS-CoV-2 sequences published by the Robert-Koch-Institut on Github. The data here is updated every hour, automatically through a Github action, so whenever new data appears in the RKI repo, you will see it here within at most an hour. Here are the first 10 lines of the dataset. IMS_ID,DATE_DRAW,SEQ_REASON,PROCESSING_DATE,SENDING_LAB_PC,SEQUENCING_LAB_PC,lineage,scorpio_call IMS-10294-CVDP-00001,2021-01-14,X,2021-01-25,40225,40225,B.1.1.297, IMS-10025-CVDP-00001,2021-01-17,N,2021-01-26,10409,10409,B.1.389, IMS-10025-CVDP-00002,2021-01-17,N,2021-01-26,10409,10409,B.1.258, IMS-10025-CVDP-00003,2021-01-17,N,2021-01-26,10409,10409,B.1.177.86, IMS-10025-CVDP-00004,2021-01-17,N,2021-01-26,10409,10409,B.1.389, IMS-10025-CVDP-00005,2021-01-18,N,2021-01-26,10409,10409,B.1.160, IMS-10025-CVDP-00006,2021-01-17,N,2021-01-26,10409,10409,B.1.1.297, IMS-10025-CVDP-00007,2021-01-18,N,2021-01-26,10409,10409,B.1.177.81, IMS-10025-CVDP-00008,2021-01-18,N,2021-01-26,10409,10409,B.1.177, IMS-10025-CVDP-00009,2021-01-18,N,2021-01-26,10409,10409,B.1.1.7,Alpha (B.1.1.7-like) IMS-10025-CVDP-00010,2021-01-17,N,2021-01-26,10409,10409,B.1.1.7,Alpha (B.1.1.7-like) IMS-10025-CVDP-00011,2021-01-17,N,2021-01-26,10409,10409,B.1.389,
Read more